Ancestry testing through genetics
I recently had a genetic analysis done to determine my ancestry - courtesy Ancestry24, theMedical Research Council, the National Health Laboratory Services and WITS University. It was a free test, and it came with an informative booklet to explain the results.
Through the African Institute for Mathematical Sciences I was given an opportunity to participate in a free Genealogical DNA test to trace my ancestry. They can do two tests for men, and one test for women. The tests trace you back through your mothers-mothers-mother or your fathers-fathers-father to the most recent ancestor with a non-coding DNA mutation.
These are the same tests they do at a crime scene, and as a paternity test. These are NOT the same tests done for diseases and allergies.
Introduction
The Nuclear Genome made up of a 50-50 mixture of DNA from our mother and our father. It is on 23 pairs of chromosomes, and adds up to about 3 billion base pairs. It is this genome that accounts for our physical differences. One of these chromosome pairs determines your sex - X-X for women, X-Y for men. The sperm carries half of the mans chromosome with it, and it statistically has a 50% chance of being an X or a Y. Thus the Y Chromosome is passed from the father to the son - there is no trace of this in women.
There is another structure inside every cell, the Mitochondria that converts energy from food into a form that cells can use, ATP. Mitochondria are found in the cells of animals, plants, algae and their common precursor, the fungi. In humans this consists of about 16,500 base pairs, and comes entirely from the mother.
If we go back another generation, to our grandparents, we are made up of 25% each of their genetic material. But there are two of them that we can trace easily - the grandfather on the father’s side, and the grandmother on the mother’s side. These two out of the four grandparents are not ‘special’ in any other way - we inherit characteristics from all four - we just have the ability to track just two.
Another generation - 2 grandparents out of 8 are ’trackable’ for men - only one out of the eight is trackable by women.
It is important to note that the mutations that occur do so at random. Most mutations will be unsuccessful, resulting in proteins that do not work. Also, to ‘stick’, these have to happen at the level of the sperm or the egg. A cosmic ray came down and struck him in the balls, as it were, and that sperm made it to the goalposts.
The people that this happened to are also not necessarily particularly special. It just happened that one of their descendants (down the male or female line, depending on whether we are talking of the mitochondrial or Y chromosome) was successful, as a mother or in warfare and survival. These changes happen on average every 19,000 years, so the gap between mutation and their descendant’s ‘success’ could be many generations.
The scientific process to sequence genes is very complicated, but is now done almost entirely by machine.
Mitochondrial DNA

For the purposes of ancestry testing, we do not use the entire genome. For Mitochondrial DNA, about 800 base pairs from two regions are used. This is done for a number of reasons - firstly, it is a lot less work to accurately sequence a (relatively) small section, and secondly this section is ‘junk’ - it does not code for any proteins, and thus plays no part in evolution. It also appears to be subject to change - the two sections are called “hypervariable regions (HVRI and HVRII)”, making it a suitable place to look for mutations.
The 800 base region Mitochondrial DNA is compared one-by-one against what is called the “Cambridge Reference Sequence”. This reference sequence is not ‘special’ at all - it just happens to be the first one done. All others are simply listed as a ‘diff’ against that one.
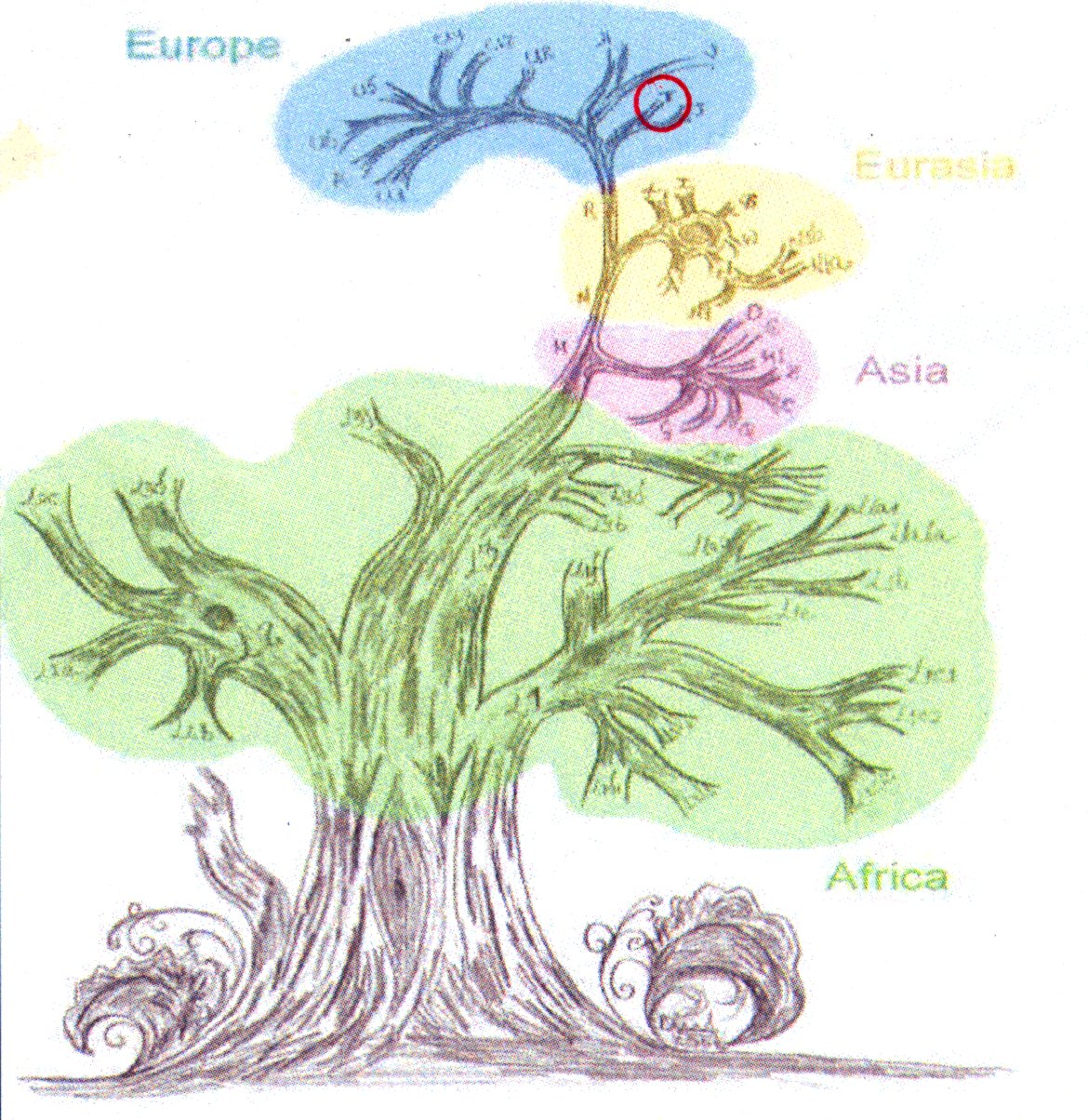
All the changes found can be mathematically tested for ‘distance’, which builds a tree. It is considered mathematically unlikely that one change will be exactly reversed by another mutation, as there is no selective reason to do so. This tree can thus be considered to represent inheritance - with the MitochondrialEve at the root of the tree. She is believed to have lived about 140,000 years ago in what is now Ethiopia, Kenya or Tanzania. Note that this is the most recent common ancestor (MRCA) of all humans via the mitochondrial DNA pathway, not the unqualified MRCA of all humanity.
The most interesting branch of the tree is that closest to Mitochondrial Eve. This is the L0 (L-nought) branch, and specifically L0d, which has high representation amonst the Khoisan. A part of Ancestry24’s goal was to sequence genes of people living in the Cape Town area and ‘catch’ some more samples in the sexy L0 branch, from the Cape Coloured population. They got some - I seem to remember about 15 or so. The DNA tree is typically represented as the root (Mitochondrial Eve, no individuals found with that exact sequence), a low-hanging branch (L0 and branches), and working up to Eurasians at the ’top’. Of course, we are all the same ‘age’ - and some african and asian branches have just as many variations, but it still Euro-centric to put us at the ’top’. Ancestry24 are just trying to get more data to populate the lesser-studied branches - sampling skew says they have many more representatives from Europe and North America. North America carries an old asian branch in the American Indians - where studies first started. Haplogroups A, B, C, D are those groups represented well in American Indians. They also have fairly good data from West Africa - via their African-Americans and the slave trade.
What does it mean to have a certain mtDNA Haplogroup ? For example, my detailed results are as follows :-
- Changes in HVR1
- 16126T-C, 16294C-T,16296C-T, 16304T-C
- Changes in HVRII
- 73A-G, 263A-G
- mtDNA Haplogroup
- T2
The Cambridge reference sequence belongs to European haplogroup H, and these represent the ’turns’ needed on the tree to ‘drive’ from H to my leaf of the tree, T2. In the larger picture, T2 is a subdivision of T, which is a subdivision of R, which is a subdivision of N, which is a subdivision of L3.
Running that backwards, the booklet that came with my results tells me that L3 is thought to have originated about 80,000 years ago, most likely in East Africa. It is associated with an exodus “Out of Africa” about 60,000 to 80,000 years ago. The daughter lineages of L3, M and N left Africa to populate the rest of the world.
European haplogroups have been extensively studied, and Bryan Sykes wrote a book called “The Seven Daughters of Eve” where he personalised the subgroups that made it to Europe. T is given to the clan of Tara, who is hypothesised to have lived about 17,000 years ago in the northwest of Italy among the hills of Tuscany and along the estuary of the river Arno. Her descendants form about 10% of modern europeans.
Y-chromosome DNA
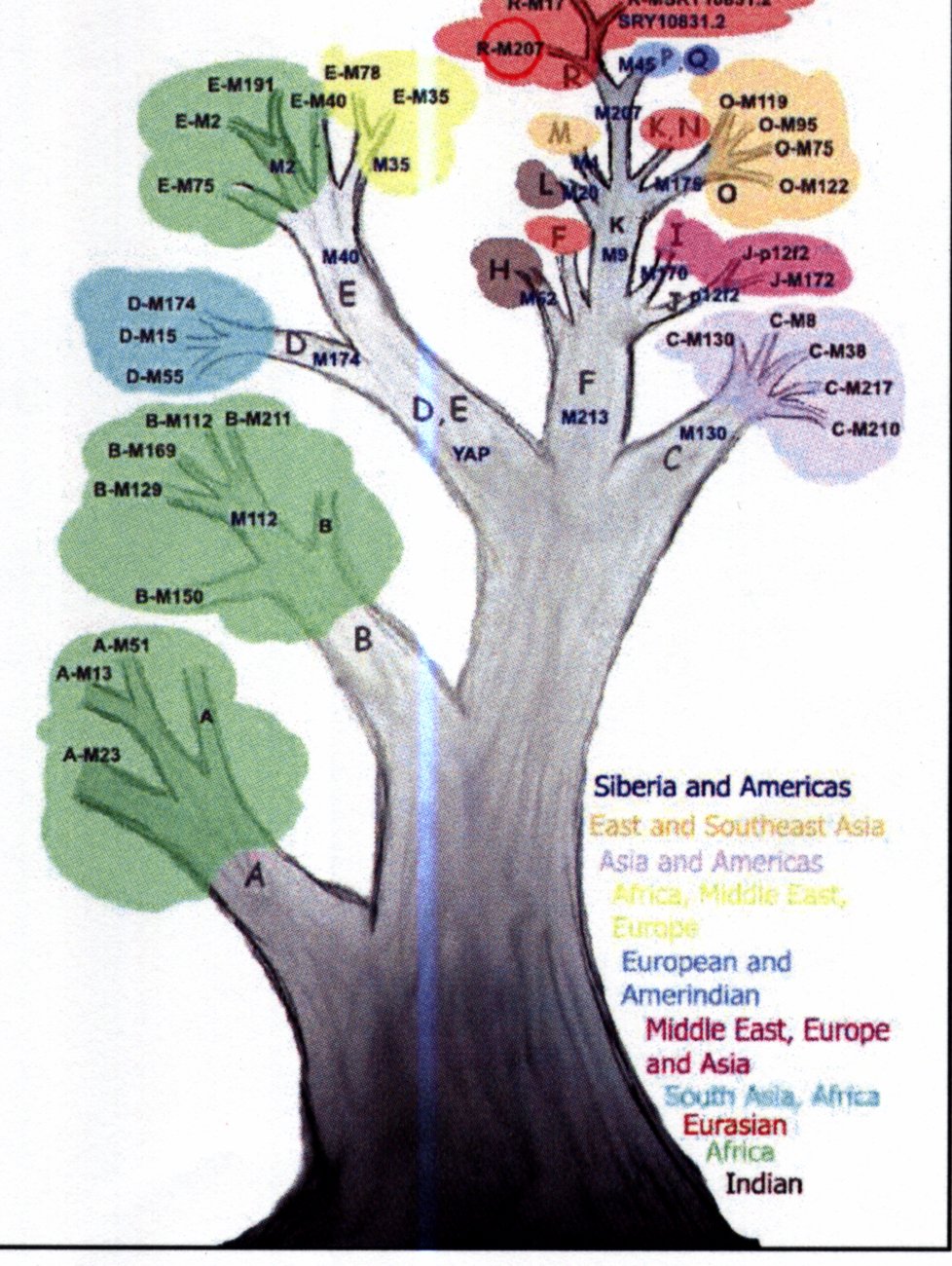
I was lucky (being a man) that I also get a Y chromosome analysis. Women share this lineage with brothers and fathers. Thus my Y chromosome analysis is identically applicable to my sisters (and brothers and father). The test done is different to the Mitochondrial DNA, and is considerably more difficult to extract, as it is a part of the nuclear DNA (3 billion base-pairs). Two types of markers are used - bi-allelic variants (two states or alleles can be found at one site on the chromosome) are used to classify me into a haplogroup. I am in Haplogroup R star - an as-yet unclassified sub-branch of R. This lettering bears no relation to the lettering of mtDNA above. The other marker used is called the STR (Short Tandem Repeats) consist of repetitive DNA elements that are tandemly repeated and are highly variable in humans. It is treated like a ‘hash’ - it will uniquely identify you but otherwise has little information on my origins.
As you can see, the STR data is useful for crime scene analysis, and close relative comparisons. I won’t be putting that information on my blog :-)
So, where does R star come from ? My booklet is a lot more vague about the origins of the Y Chromosome haplogroups, talking of scattering, movement, displacement. This makes sense. Men can father more children, and in war, particularly centuries-old forgotten internecine wars in africa, the victors would have killed the males and taken the females as wives. Such is life. A man may father many sons, and lose them all as quickly. The female line is steadier.
The mathematics of tree-building does not care about mens habits in warfare- it unambiguously records the successive mutations in the Y chromosome. R is a sub-group of P which is a sub-group ofK which is a sub-group of F. F branched from an untraceable parent, and F’s peers are A, B, C, D, and E.
Haplogroups A thru E are variously represented in Southern Africa, with the Khoisan in A3b1. B2b is also found in the Khoisan and Pygmy populations of central africa. E3a most likely spread with the Bantu expansion, and is now the most common haplogroup in sub-Saharan Africa. Way-to-go E3a!
But .. hats off to F. A super-haplogroup, it and its sub-lineages contain more than 90% of the worlds male population. It possibly originated some time between 45,000 and 80,000 years ago, somewhere in North Africa or the Middle East. Some believe it travelled out of Africa on the first migration, some place it at the second migration.
K cannot be geographically placed well, though it is an old lineage. P is believed to have arisen in Siberia, Kazakhstanor Uzbekistan 35,000 to 40,000 years ago. R is thought to have originated somewhere in Northwest Asia around 30,000 years ago, and is primarily represented in Europe and West Eurasia.
So, the trail is lost somewhere in Europe, with movements occuring too quickly to be tracked by genetic mutations.
My thanks to Dr. H Soodyall and her team at the Dept. of Human Genetics for all their hard work bringing this to us.
Postscipt - commercial DNA testing
I have not used any of these sites, but here is a review site that was sent to me that compares the different approaches of online DNA databases.
Not mentioned in the review - but do be aware that your personal identification can be stored on these sites, which in turn can be hacked.